Co-authorship in Nature Genetics with SickKids

Overview #
I spent two co-op work terms doing bioinformatics research at The Center for Applied Genomics (TCAG) at the Hospital for Sick Children (SickKids), where I had the opportunity to work on studying how missense variants affected protein-protein interactions.
While there, I had the opportunity to contribute to a paper covering the genomic architecture of cerebral palsy. My contributions involved simulating PPI between CP-affected proteins using AlphaFold-Multimer in an attempt to identify any notable differences between wildtype and missense interactions, as well creating a supplemental figure.
Here’s a quote from the paper describing the research I worked on: 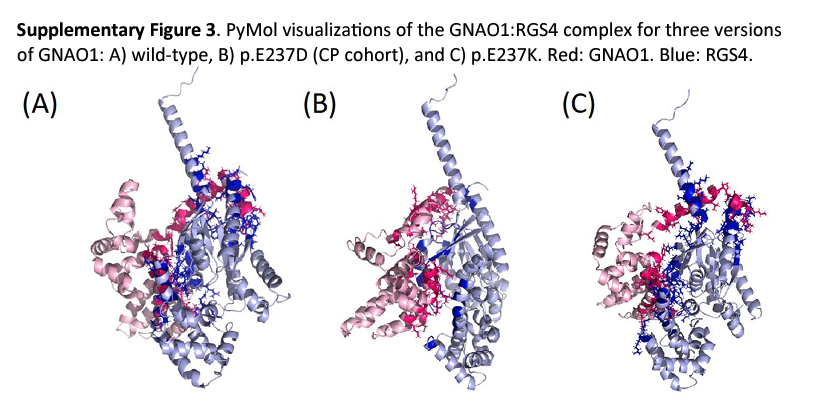
To gain preliminary functional insight into potential molecular mechanisms of missense variants detected in the CP cohort, we used in silico approaches to investigate two potential mechanisms: disruption of protein–protein interactions34 and disruption of phosphorylation-mediated signaling. We investigated three selected de novo P/LP missense variants using AlphaFold-Multimer to assess whether their mechanisms could disrupt protein–protein interactions (p.A334T in TUBA1A, p.D132A in EXOSC3 and p.E237D in GNAO1). These variants were selected based on previous evidence that missense variants in their respective proteins may disrupt protein–protein interactions. TUBA1A and EXOSC3 were not predicted by AlphaFold-Multimer to be involved in the interface with their putative interacting protein(s) (Supplementary Table 8, sheet 1). However, the GNAO1 variant, p.E237D, was predicted by AlphaFold-Multimer to be involved in the interface with its regulator RGS4. It is interesting that a de novo variant at the same position, but causing a different amino acid change (p.E237K), has previously been implicated in CP-like features40. We modeled both missense variants in AlphaFold-Multimer to assess the GNAO1–RGS4 interaction. Despite the different amino acid characteristics of the two missense variants (aspartate versus lysine), they were predicted to have similar effects on the GNAO1–RGS4 interface (Supplementary Table 8, sheet 2 and Supplementary Fig. 3). Furthermore, the p.E237D structure was predicted to be more similar to the p.E237K structure (distance = 0.28) than to the wild-type structure (distance = 0.45).
Skills #
- Collaborative research: Before my co-op, I had no idea how much work went into the development of a research paper. Countless meetings, conversations, and SLURM jobs.
- Independent research and discovery: As one of the only members of the lab working on an interactomics, I laid much of the groundwork for the project myself. Luckily, I had great mentors in Dr. Brett Trost and Dr. Bank Eungchan.
- AlphaFold-Multimer: AF-M is an incredible tool (especially in conjunction with a tool like PyMol), solving some of the most elusive problems in the world of computational molecular biology. Getting to work with it every day was incredibly illuminating.
Key Takeaways #
- There’s a lot left to do: While it may seem like AlphaFold has answered every question we have, it’s opened up even more. Can we design algorithms that predict the effects of structure disrupting/altering point mutations? Can we have them design proteins for us? There’s so much opportunity available.
- Research takes a long time: I first learned the paper would be getting published in mid-2022. Publication occurred nearly a year later. No complaints, obviously, but it’s a strong reminder of the checks and balances required to put out quality research.